Factorial Randomized Block Design along with LSD test in R
The post explains how to get FRBD ANOVA, Interpretation of ANOVA, R-square, Normality assumption testing, Least significant difference (LSD) test using doebioresearch package in RStudio (Reading time 12 min.)
R-script
#This line will load doebioresearch package
library(doebioresearch)output
print(output)
sink()
Output and interpretation
Analysis of Variance Table
Response: dependent.var
Df Sum Sq Mean Sq F value Pr(>F)
replicationvector 2 61.06 30.53 1.4112 0.276494
fact.A 1 320.47 320.47 14.8123 0.001772 **
fact.B 1 56.12 56.12 2.5939 0.129584
fact.C 1 154.53 154.53 7.1426 0.018208 *
fact.A:fact.B 1 272.70 272.70 12.6043 0.003199 **
fact.A:fact.C 1 0.22 0.22 0.0102 0.921034
fact.B:fact.C 1 3.60 3.60 0.1666 0.689355
fact.A:fact.B:fact.C 1 3.01 3.01 0.1391 0.714722
Residuals 14 302.90 21.64
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
The ANOVA shows Fact.A (Paddy variety), Fact.C (Irrigation schedule) and Fact.A*Fact.B (Paddy variety*Plant protection schedule) were significant. We will ignore the the LSD results of rest of the combination of the factors and interaction.
[1] "R Square 0.742"
0.742 i.e. 74.20% of variation present in the dependent variable is explained by the model.
[1] "SEm of A: 1.343 , SEd of A: 1.899 , SEm of B: 1.343 , SEd of B 1.899 , SEm of C: 1.343 , SEd of C: 1.899 , SEm of AB: 1.899 , SEd of AB: 2.685 , SEm of AC: 1.899 , SEd of AC: 2.685 , SEm of BC: 1.899 , SEd of BC: 2.685 , SEm of ABC: 2.685 , SEd of ABC: 3.798"
Shapiro-Wilk normality test
data: model$residuals
W = 0.98223, p-value = 0.9332
[1] "Normality assumption is not violated"
[1] "The means of one or more levels of factor A are not same, so go for multiple comparison test"
MSerror Df Mean CV t.value LSD
21.63548 14 119.6458 3.887636 2.144787 4.072787
dependent.var groups
v0 123.3000 a
v1 115.9917 b
Paddy variety vo (ADT-31) gives highest yield which is significantly different from v1 (Vaghai)
[1] "The means of one or more levels of factor C are not same, so go for multiple comparison test"
MSerror Df Mean CV t.value LSD
21.63548 14 119.6458 3.887636 2.144787 4.072787
dependent.var groups
w1 122.1833 a
w0 117.1083 b
The irrigation schedule new practice gives highest yield which is significantly different from the local practice
[1] "The means of levels of interaction between A and B factors are not same, so go for multiple comparison test"
MSerror Df Mean CV t.value LSD
21.63548 14 119.6458 3.887636 2.144787 5.759791
dependent.var groups
v0:p0 128.2000 a
v0:p1 118.4000 b
v1:p1 117.8333 b
v1:p0 114.1500 b
The combination v0p0 (ADT-31*old practice) has highest yield which is significantly different from the rest of three combinations.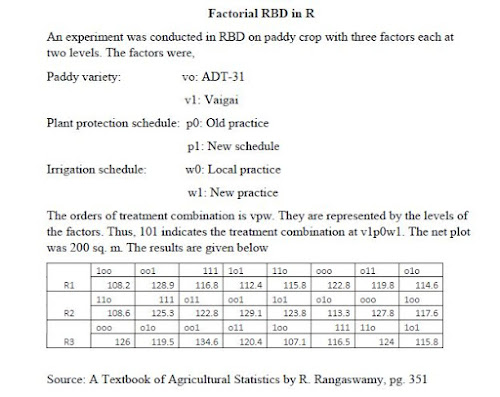

Comments
Please share the no of treatments and replication of CRD design. If possible I will share excel tool for that.
Can you please make a video on splitplot and CRD using "doebioresearch"
https://www.youtube.com/watch?v=j5FRTZDWDlA&t=32s
For CRD you can refer example given in package
Regards
RAAJ