RCBD analysis in R along with LSD and DNMRT test
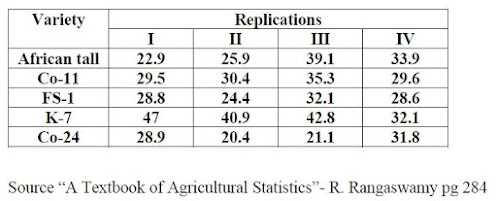
The blog explains step by step RCBD analysis along with LSD/DNMRT test using R and interpretation of analysis (Reading time 12 min.) Problem statement An experiment was conducted in RCBD to study the comparative performance of fodder sorghum lines under rain fed conditions. Data is furnished below. Are all lines same? If not carry out LSD test and Duncan test to compare the lines. Click here to get the data Arrange the data in Excel as shown below Replication Variety Yield 1 African tall 22.9 1 Co-11 29.5 1 FS-1 28.8 1 K-7 47 1 Co-24 28.9 2 African tall 25.9 2 Co-11 30.4 2 FS-1 24.4 2 K-7 40.9 2 Co-24 20.4 3 African tall 39.1 3 Co-11 35.3 3 FS-1 32.1 3 K-7 42.8 3 Co-24 21.1 4 African tall 33.9 4 Co-11 29.6 4 FS-1 28.6 4 K-7 3

