Split plot analysis in R using doebioresearch package
The blog explains split plot analysis in R along with assumption testing and LSD test for mean comparison (Reading time 9 min)
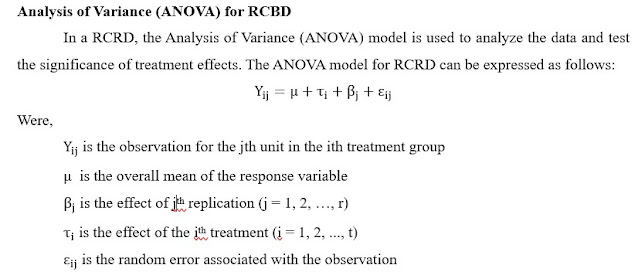
Click on the image to expand
Replication: 4
Main-plot: Variety levels 3
Sub-plot: Nitrogen levels 3
Dependent variable: Yield
Source of problem: https://fliphtml5.com/waov/gjll/basic
Data: excel sheet
R-script: script
#Upload excel file into RStudio
library(readxl)
SP <- read_excel("D:/Youtube/Design of
Experiments/Split plot/SP.xlsx")
View(SP)
#You can upload the data by clicking on import data set-> From Excel -> Browse -> Select your file-> #Import
#You can upload the data by clicking on import data set-> From Excel -> Browse -> Select your file-> #Import
#Load doebioresearch package
library(doebioresearch)
#Store split plot analysis in variable named output along with LSD test
output<-splitplot(SP[4],SP$Replication,SP$Variety,SP$Nitrogen,1)
#Store output in document outside RStudio in word file
sink("output.doc")
print(output)
sink()
Output from R:
Analysis of Variance Table Response: dependent.var Df Sum Sq Mean Sq F value Pr(>F) block 3 190.083 63.361 2.1836 0.190936 main.plot 2 90.487 45.243 1.5592 0.284900 Ea 6 174.100 29.017 sub.plot 2 92.435 46.217 10.2287 0.001078 ** main.plot:sub.plot 4 9.533 2.383 0.5275 0.717003 Eb 18 81.332 4.518 --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 The p-value of the subplot is <0.05, so its significant. The means of all the three nitrogen levels are not same so go for LSD test. The main plot and interaction is non significant so ignore their LSD test result and all of their levels are same.
[1] "CV(a): 30.348 , CV(b) : 11.976" The main plot C.V. is high and out of general limit which will need a suitable justification. The C.V. for the sub plot is within a limit (less than 12-15%).
[1] "R Square 0.873" 0.873 i.e. 87.3% variation present in the dependent variable yield is explained by the model. Shapiro-Wilk normality test data: model$residuals W = 0.97257, p-value = 0.5 [1] "Normality assumption is not violated"
$Yield[[1]][[6]] [1] "All the main plot factor level means are same so dont go for any multiple comparison test" [1] "The means of one or more levels of sub plot factor are not same, so go for multiple comparison test" MSerror Df Mean CV t.value LSD 4.518426 18 17.75 11.97554 2.100922 1.823173 dependent.var groups n3 19.70833 a n2 17.75833 b n1 15.78333 c The LSD test reveals that 60 kg/ha (n3) gives highest yield which is on par with/statistically different from 30 kg/ha (n2) and 0 kg/ha (n1).
Happy Learning!
Bored of reading? Tune to video!
Hope you found this blog useful. If have any query or suggestions please do post it below.
If you are using "doebioresearch" for analysis. You can cite it as:
Popat, R and Banakara, K. (2020). doebioresearch: Analysis of Design of Experiments for Biological Research. R package version 0.1.0
To learn more about Agricultural Statistics follow my youtube channel
Topics you might be interested in:
Degrees of freedom
Principles of designs of experiments-II: Randomization
Principles of design of experiments-III: Local control
Principles of design of experiments-III: Local control


Comments
Very good your explanations about the application of this library. To date it is one of the simplest I have found for this type of analysis. I am working recently with R and I have a very similar design but with multiple responses. I want to graph the varieties and treatments, but keep it in a single figure, but with the three separate graphs. First treatment and the three varieties, second and three varieties, etc. Is it possible? Could you help me? Thank you
Thanks for the compliments. I would not be able to help you in context of graph as I know little about visualization in R.
Regards
RAAJ
$Yield[[1]][[6]]: what does it mean? Coudl you please explain it?
Hope you got it
Regards
RAAJ
Please, I want to thak you for your videos.
I like the video on split plot.
My question is how do I code to have let's say "Plant stand harvested" as a covariate for "yield"?